Detection of RNA secondary structure motifs using computational vision approach
Final project by
Nir Dromi
Introduction
the RNA molecules has essential roles in large variety of biological processes.
the understanding of the secondary structure of a RNA molecule, is importent to understand it function in the biological processes.
there are some computational tools that automatically finds the secondary structure of a RNA molecule, given it nucleotides sequence.
in order to find RNA molecules with some interesting attributes, bioinformatic researchers needs to check a very large number of RNA secondary
structure pictures.
in this project we give a tool that automatically examine a secondary structure picture, and write down it structure's main motifs in
a formal way.
Approach and Method
we developed a tool that takes a secondary strucure of RNA molecule, represent it as a non-directed graph,
and analyze the matrix describing this graph as if it was a picture.
the goal is to understand from the picture (the matrix) what are the main motifs in the RNA structure.
the RNA structural motifs recognition is done by a series of actions:
1) taking a RNA sequence as an input. for example:
GGCAGUACCAAGUCGCGAAAGCGAUGGCCUUGCAAAGGGUAUGGUAAUAAGCUGCC
2) compute the secondary structure of this molecule, by Mfold Web Server
for example:
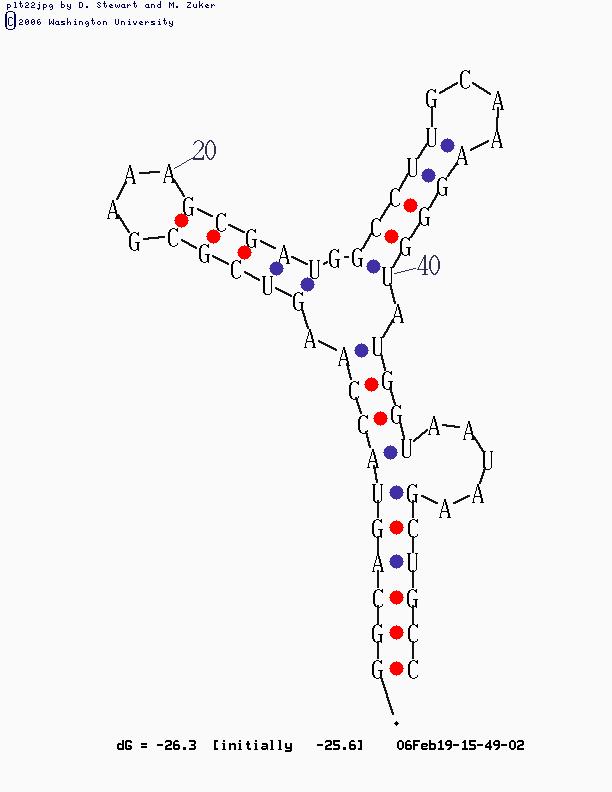
3) represent the the structure by a non-directed graph (the representation method is shown in the full project report).
for example:

4) identification of structural motifs by operating image processing tools on the matrix's picture.
there are 3 main motifs in the RNA molecular structure : hair-pin, bulge and internal-loop (shown in the full project report).
my approach deal with the problem in two steps:
a) detecting the motifs in the picture.
b) identifing the type of each motif.
my original approach to the problem (of identifing the type of each motif),
was to solve it with the method of relaxation labeling, where:
the Objects are the motifs in the picture
the labels are the motif types (hair-pin, bulge and internal-loop)
the compatibility is the set of constraint that the motifs has on each other, depending on it location on the matrix.
while creating the compatibility rules, it has been clear that it is possible to identify the types in a deterministic way,
without the need of relaxation labeling, but only with the use of four attributes of the matrix.
these attributes gives for every motif it type (the attributes are shown in the full project report).
the following is an example for the results of the algorithm on the matrix given above:
motif 1: up: (6, 51) down: (7, 45) type: BULGE
Motif 2: up: (10, 42) down: (12, 25) type: INTERNAL
Motif 3: up: (16, 21) down: (0, 0) type: HAIRPIN
Motif 4: up: (31, 36) down: (0, 0) type: HAIRPIN
Results
the algorithem develeped for detecting the 3 main motifs (hair-pin, bulge and internal-loop) is determenistic,
therefore it detects these motifs whenever they appear.
for example, in the structure below, the four motifs have been detected:
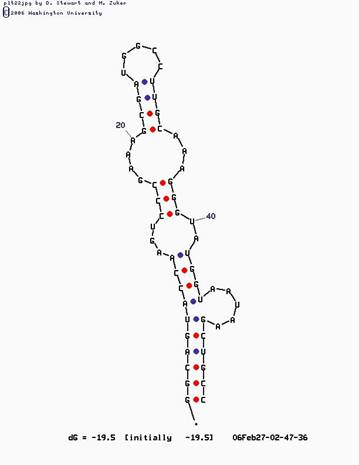

the results of the algorithm:
motif 1: up: (6, 51) down: (7, 45) type: BULGE
Motif 2: up: (10, 42) down: (14, 39) type: INTERNAL
Motif 3: up: (16, 37) down: (21, 33) type: INTERNAL
Motif 4: up: (24, 30) down: (0, 0) type: HAIRPIN
Conclusions
it is possible to develop a determenistic algorithm that takes the matrix that represent the RNA structure,
and learn from it what are the main motifs in the structure.
the original approach to the problem was to solve it with the method of relaxation labeling,
but as the work went on i figured out that because of the assumptions that i could make on this case,
it was possible to develop a determistic algorithm, that gives much better results than the
relaxation labeling method.
Additional Information
References
[1]M. Zuker, "Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction, "Nucleic Acids Res., Vol.31, pp.3406-3415,2003
Mfold Web Server
[2] I.L. Hofacker, "Vienna RNA Secondary Structure Server," Nucleic Acids Res., Vol.31, pp.3429-3431,2003.
Vienna RNA Secondary Structure Server
[3] M.S. Waterman, "Secondary Structure of single-Stranded Nucleic Acids,"Studies on foundations and combinatorics, Advances in nathematics supplementary studies, Academic Press N.Y., Vol.1, pp.167-212, 1978.
[4] T. Jiang G.H. Lin, B. Ma, and K. Zang, "A General Edit Distance Between RNA structures, "J. Comput. Biol., Vol.9, pp.371-388, 2002.
[5] D. Barash, "Second Eigenvalue of the Laplacian Matrix for Predicting RNA Conformational Switch by Mutation," Bioinformatics, 10.1093/bioinformatics/bth157,2004.
[6] M. Fiedler, "Algebric Connectivity of Graphs," Czechoslovak Math. J., Vol.23,pp.298-305,1973.
[7] S.Y. Le, R. Nussinov, and J.V. Maizel, "Tree Graphs of RNA Secondary Structures and Their Comparisons", Comput.Biosci. Vol.6,pp.309-318,1990.
[8] E. Rivas, S.R. Eddy, "A dynamic programming algorithm for RNA structure prediction including pseudoknots",J. of Molecular Biology. Vol 285, pp2053-2068, 1999.



