IndelsRNAmute source code and executable jar file:
How to use IndelsRNAmute
The current vertion of IndelsRNAmute is available only for Windows and was tested on a standard PC with Windows 10.Before using IndelsRNAmute please install the following two programs for Windows and add these programs to Windows system PATH:
- Vienna RNA package 2 from: https://www.tbi.univie.ac.at/RNA/#download
- ImageMagic from: https://imagemagick.org/script/download.php
After downloading IndelsRNAmute:
1) Extract the file "IndelsRNAmute.zip".
2) Enter the folder "IndelsRNAmute".
3) Run "IndelsRNAmute.jar" file.
The IndelsRNAmute software uses code developed and compiled in C: bin/Multi_Mut.exe. In case the Multi_Mut.exe does not work on your computer, please compile C code available in src/Multi_Mut.c. The code was compiled and tested in Visual Studio 2015.
A short tutorial
Input screen with example parameters:
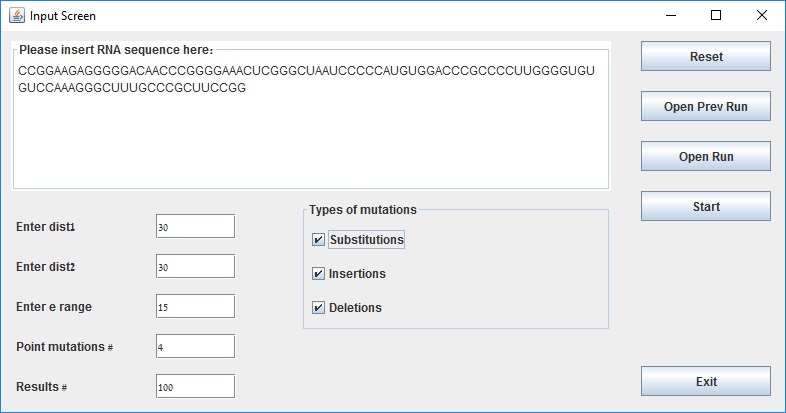
Input screen includes:
1) RNA sequence field - the maximum sequence length allowed in our application is 1000 bases.
2) dist 1 - this distance parameter is used for filtering suboptimal solutions that are close to the optimal solution. The suggested value to use is around 30% of the RNA sequence length.
3) dist 2 - this distance parameter is used for filtering suboptimal solutions that are close to each other. The suggested value to use is around 30% ofthe RNA sequence length.
4) e range - this energy parameter is used in the RNAsubopt program to calculate the suboptimal structures within a range of kcals/mol of the mfe. The suggested value is around 15% of the RNA sequence length.
5) Point mutations # - number of allowed point mutations in RNA (one M-point mutation set).
6) Results # - number of M-point mutations in the output.
7) Type of mutations (SUM, INS, DEL) - the user may choose to allow insertions, deletions and substitutions in the M-point mutation set.
8) Open Prev Run - The application saves the results in a file, allowing to open previous runs without running the application again.
9) Open Run - The user may save the results and insert them later in the GUI.
10) Start - To start the calculation press this button. After the calculation terminates, the Results screen will be displayed.
Results screen (for example paremeters):
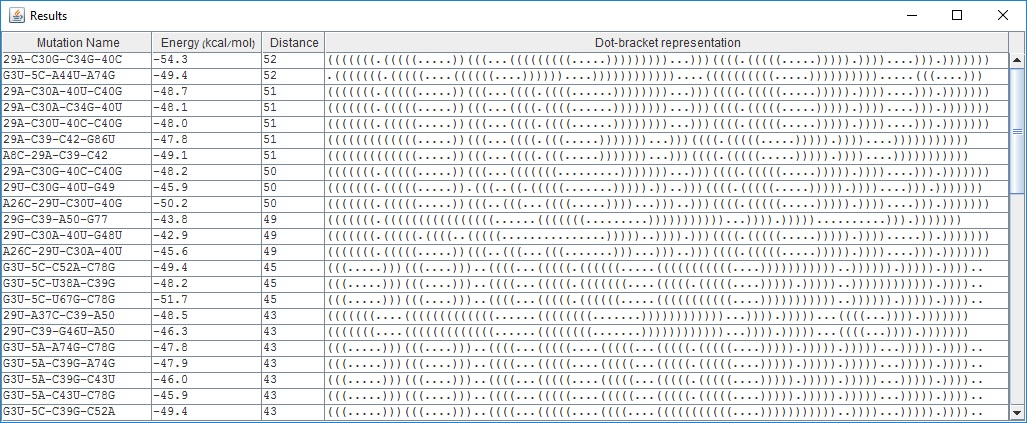
The Results screen lists up to 100 4-point mutations, sorted by distance of their structure from the wild type structure. The most deleterious mutations are listed first. Each row in the table includes the mutation name, free energy, distance from wildtype RNA and the dot-bracket representation of its structure.
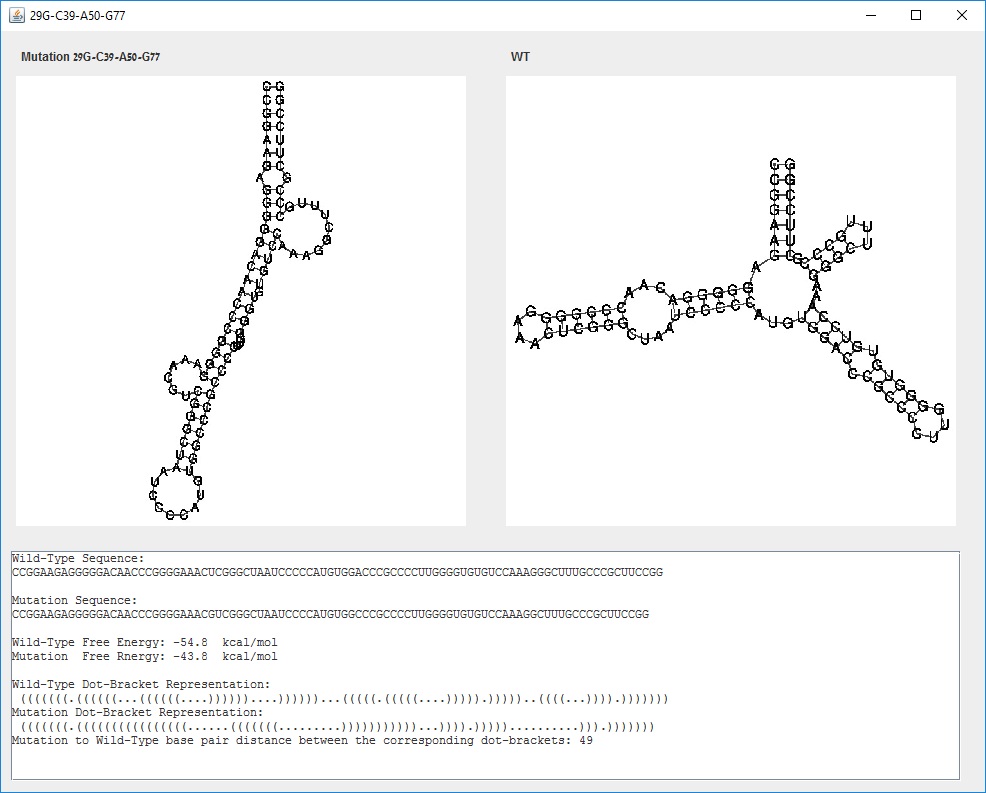
The user may further investigate a given mutation by pressing on some row in the table to see more information about a specific mutation. For example, selecting the mutation 29G-C39-A50-G77 will open the Mutation screen.
Mutation screen for selected mutation: